*Paraphelidium* transcriptome: some code
February 11th, 2019 by xgrau
This is a quick post to highlight the publication of Global transcriptome analysis of the aphelid Paraphelidium tribonemae supports the phagotrophic origin of fungi by Guifré Torruella, Purificación López-García (Université Paris-Sud) et al., in Communications Biology.
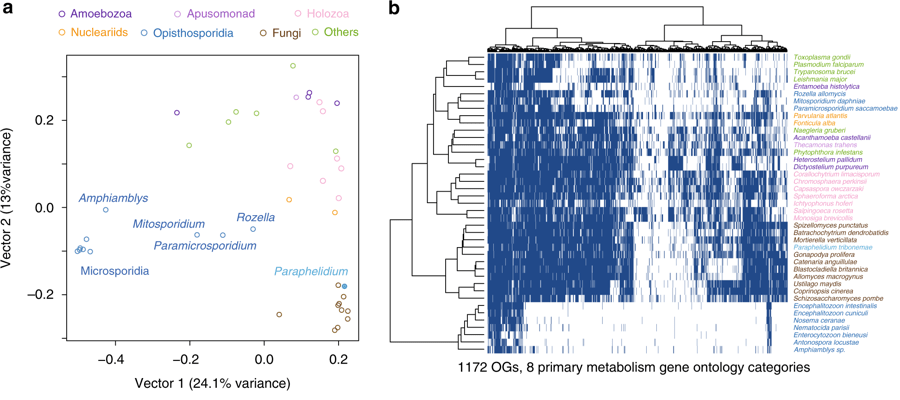
In parallel, we released the R code we used to analyse the functional profile of its gene content, which you can find it in this Github repository. We used the absence/presence profile of a set of genes linked to primary metabolism in the genomes (or transcriptomes) of unicellular eukaryotes to highlight similarities between the gene content of fungi and Paraphelidium.
Thus, it can assist in the production of plots such as the following (Figure 3 in the paper):
The script dimreduction-heatmaps_Torruella_et_al_2018.R takes as input a tab-separated table where rows are genes/orthogroups and columns are various species. You can load any table you wish by editing these lines, right at the beginning of the script:
input = "primary_transposed.txt" # assumes a wide-style matrix with headers, rows=genes/OGs & cols=sps
mi = read.table(input,header=T,sep="\t")
Our results highlight the similarities between the rich primary metabolism of Paraphelidium and ‘canonical’ fungi. Thus, unlike other early-branching fungal allies such as Rozella, Paraphelidium does not have a simplified metabolism. This is consistent with what is actually the most interesting result of the paper (in my opinion): the phylogenomic analysis of Paraphelidium consolidates its position as sister-group to fungi and breaks up its association with Rozella and microsporidians. This has important implications regarding the lifestyle of the ancestral fungi, which was likely phagotrophic (instead of osmotrophic).
But you should definetely read the paper to get the whole picture ;)